This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
RNAi
RNAi or RNA interference is a system within cells that regulates gene expression. There are two small molecules (microRNA and small interfering RNA) which can bind to RNA and either up-regulate or down-regulate their activity. RNAi plays a role in defending cells from viral attacts, in development, and in regulating gene expression. RNAi has proven to be very useful, because it has been implemented in experiments to knock down gene expression. By knocking out a specific gene, one can observe the phenotype of an organism and determine the effect that the lack of the gene has on development/function of the organism. By observing the result of a lack of the gene, researchers can further understand the function of the gene.
Phenobank is an RNAi database that allows you to search for experiments that implicated RNAi in their research. From these studies, you can observe the phenotypic effect of knocking out gene function on the model organism, which in this database's case is C. elegans or the worm.
As there is no hcrtr1 homolog in C. elegans or Drosophila, I instead searched Phenobank for a domain rather than the specific gene. The only known protein domain in hcrtr1 is the 7 transmembrane G-protein coupled receptor. I was able to search for this domain in C. elegans, but the search returned 199 hits. After reviewing around 20 of them, most of the RNAi experiments done to the various genes found in this domain resulted in wild-type. However, the experiments on genes that did have phenotypic effects returned such results as reduced progeny, slower development, and embryonic lethality.
Phenobank is an RNAi database that allows you to search for experiments that implicated RNAi in their research. From these studies, you can observe the phenotypic effect of knocking out gene function on the model organism, which in this database's case is C. elegans or the worm.
As there is no hcrtr1 homolog in C. elegans or Drosophila, I instead searched Phenobank for a domain rather than the specific gene. The only known protein domain in hcrtr1 is the 7 transmembrane G-protein coupled receptor. I was able to search for this domain in C. elegans, but the search returned 199 hits. After reviewing around 20 of them, most of the RNAi experiments done to the various genes found in this domain resulted in wild-type. However, the experiments on genes that did have phenotypic effects returned such results as reduced progeny, slower development, and embryonic lethality.
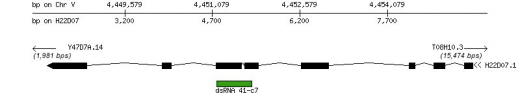
Figure 1: This depicts the summary of the RNAi information on gene H22D07.1. This particular gene is a G-protein coupled receptor found in C. elegans. It is located on chromosome 5. The results of the RNAi experiment had no effect on phenotype though (resulted in wild-type). Courtesy of Phenobank.
Analysis
While there were no RNAi experiments that existed for hcrtr1, the other 7 transmembrane receptors (many of which were G-protein coupled receptor proteins) had many RNAi experimental phenotypes. This makes sense because this receptor family is a very large family of proteins, and unsurprisingly has homologs in C. elegans. In addition, of the phenotypes described in the experiments, none had functions that resembled what hcrtr1 does in the human body. This also does not come as a surprise, because there are so many G-protein coupled receptor genes and they do not all have the same function. However, it is interesting to note that the phenotypic effects that these knockouts had were very severe. Therefore, it seems evident that these receptor proteins play important roles in many biological functions and in many organisms.
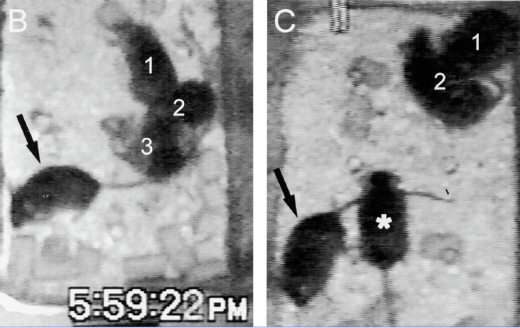
While no mutant of hcrtr1 exists in C. elegans, researchers have made knockout mice which lack the hypocretin ligand. These mice exhibit narcoleptic phenotypes such as immediately entering deep sleep. As seen in figure B below, the mouse with the arrow has fallen to his side and the other 3 mice are somewhat fuzzy because they are moving. In figure C, the two mice in the lower left have fallen asleep at odd angles, and the upper two mice are quietly sleeping in the corner. This is a similar phenotype to narcoleptic humans which enter deep REM sleep immediately.
While no mutant of hcrtr1 exists in C. elegans, researchers have made knockout mice which lack the hypocretin ligand. These mice exhibit narcoleptic phenotypes such as immediately entering deep sleep. As seen in figure B below, the mouse with the arrow has fallen to his side and the other 3 mice are somewhat fuzzy because they are moving. In figure C, the two mice in the lower left have fallen asleep at odd angles, and the upper two mice are quietly sleeping in the corner. This is a similar phenotype to narcoleptic humans which enter deep REM sleep immediately.
Figure 1: Depiction of hypocretin (orexin) knockout mice: similar to narcoleptic phenotypes. Courtesy of Chemelli et al.
References:
(1) Phenobank
(2) Chemelli RM et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999 Aug 20;98(4):409-12
(2) Chemelli RM et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999 Aug 20;98(4):409-12
Eric Suchy, Email: [email protected], last updated: May 15, 2010