This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
DNA Motifs
Motifs
This page is concerned with finding and characterizing DNA motifs that are important for the hcrtr1 gene function. A DNA motif is a widespread sequence pattern of nucleotides or amino acids suspected to have biological significance such as structure or function. I searched for different DNA motifs on the Motif website. Websites use specific algorithms to detect motifs in different genes.
I used the full FASTA format DNA sequence of hcrtr1 to search for motifs with a cutoff score of 85. I received 94 motifs, so I conducted another search with a cutoff score of 90. This search returned 30 results. Of those 30 motifs, I felt that the following were the most significant and possibly are related to the hypocretin receptor function:
I used the full FASTA format DNA sequence of hcrtr1 to search for motifs with a cutoff score of 85. I received 94 motifs, so I conducted another search with a cutoff score of 90. This search returned 30 results. Of those 30 motifs, I felt that the following were the most significant and possibly are related to the hypocretin receptor function:
- Motif 1: HSF (heat shock factor) Drosophila : this motif had the closest match with the hcrtr1 gene
- Motif 2: ADR1 (alcohol dehydrogenase gene regulator 1) : this motif seemed interesting because alcohol is known to be a depressant, which causes sleep; may have similar function with the hypocretin receptor
- Motif 3: STRE (stress-response element) : this may be important in gene function because sleep might be a response to stress
- Motif 4,5: GATA - 1,2 (GATA-binding factors) : the GATA binding factors are transcription factors binding to a receptor which is similar to the hypocretins binding to the hypocretin receptor; they might have similar functions
STAMP
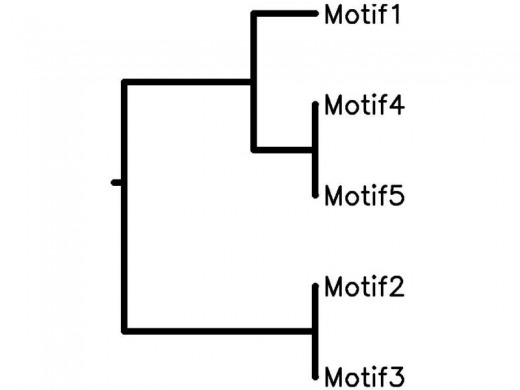
After finding motifs that seemed like they had similar functions to hcrtr1, I input them into a program called STAMP which is a website that allows you to compare alignment, similarity, and database matching for DNA motifs. The results can be seen below. The results show motifs 1, 4, and 5 (which were HSF and two GATA motifs) were closely related, and that motifs 2 and 3 (ADR1 and STRE) were closely related. The motif tree is shown below.
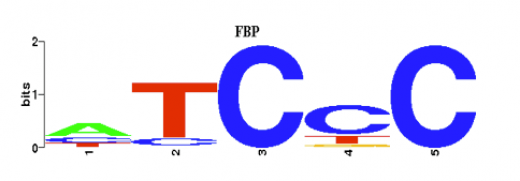
The STAMP website also allows you to view the multiple alignment of the different motifs. This figure can be seen below. It is a sequence of the compared bases between the motifs. The larger the letter is, the more likely that base is in that particular location. From the figure, we can predict that the probability of the 3rd and 5th nucleotide being C is very high. However, the usefulness of this figure in our cause is unclear, unless we were planning on engineering some changes in the base pair sequence.
Analysis
The motif website returned some interesting results. The alcohol-dehydrogenase gene regulator and the stress response element seem like promising hits because of the implications they could have in discovering the function of hcrtr1 in narcolepsy. The fact that they are closely related on the tree encourages this idea as well. Further research could go into finding any relationships between the hypocretin receptor and these motifs.
References:
Eric Suchy, Email: [email protected], last updated: May 15, 2010