This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
Homology
Genetic homology of genes and proteins refers to the similarity of sequences between various species. In some cases, similar genetic sequences arise from common ancestry. If common ancestry is not the case, then there may be other reasons for the sequence similarity including chance. Gene homology is has important implications in experiments. If a particular gene or genetic sequence is conserved in several species, then researchers can perform genetic experiments on those conserved regions in different organisms. Homologene is an online database that is used to detect homology among genes of various different organisms.
I found homologs of the human hypocretin receptor 1 protein by conducting a general search for the gene in HomoloGene. There are homologous genes in several model organisms. I then compared each homologous sequence to the human sequence using BLAST (basic local alignment search tool). Each BLAST search returned an identity percentage of the two sequences, and an expectation value (E-value). The lower the E-value, the more significant the information is based on an algorithm the program uses. Here is a list of the organisms and the accession numbers, the identity percentage, and the E-values of their homologous genes:
I found homologs of the human hypocretin receptor 1 protein by conducting a general search for the gene in HomoloGene. There are homologous genes in several model organisms. I then compared each homologous sequence to the human sequence using BLAST (basic local alignment search tool). Each BLAST search returned an identity percentage of the two sequences, and an expectation value (E-value). The lower the E-value, the more significant the information is based on an algorithm the program uses. Here is a list of the organisms and the accession numbers, the identity percentage, and the E-values of their homologous genes:
1. Pan troglodytes (Chimpanzee)
Name: Hypocretin (orexin) receptor 1
Gene Accession Number: XM_001159598
Length: 425 amino acids
FASTA sequence
BLAST identity percentage: 94%
E-value: 0.0
2. Canis lupus familiaris (Dog)
Name: Hypocretin (orexin) receptor 1
Gene Accession Number: XM_544446
Length: 502 amino acids
FASTA sequence
BLAST identity percentage: 92%
E-value: 0.0
3. Bos taurus (Cattle)
Name: Hypocretin (orexin) receptor 1
Gene Accession Number: NM_001048182
Length: 425 amino acids
FASTA sequence
BLAST identity percentage: 95%
E-value: 0.0
4. Mus musculus (Mouse)
Name: Hypocretin (orexin) receptor 1
Gene Accession Number: NM_198959
Length: 416 amino acids
FASTA sequence
BLAST identity percentage: 91%
E-value: 0.0
5. Rattus norvegicus (Rat)
Name: Hypocretin (orexin) receptor 1
Gene Accession Number: NM_013064
Length: 416 amino acids
FASTA sequence
BLAST identity percentage: 91%
E-value: 0.0
Homologs in C. elegans
There was no C. elegans homolog for hcrtr1, however the hypocretin receptor 2 (hcrtr2) did have a homologous gene in worms. The gene name is npr-14 of the neuropeptipde receptor family with the sequence name W05B5.2. It can be found on the first chromosome (Wormbank). However, no information as of yet has discovered a correlation between the hypocretin receptor 2 and narcolepsy.
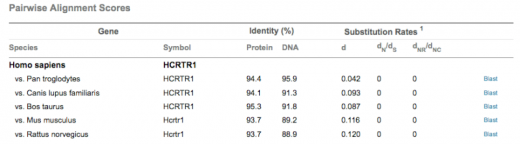
Clicking on the graph above enlarges it (or visit the link). The graph is a summary of the alignment scores of hcrtr1 between different species compared with humans. As you can see, the identity percentages from above correlate to the pairwise alignment scores.
Analysis
After reviewing the results, it appears the hcrtr1 gene is very highly conserved in these different species. The highest pairwise alignment scores are found in chimpanzees which makes sense because they are the closest human relative. Many experiments have been conducted with narcoleptic mice and rats. Both of these species have very high homology scores. In addition, the first species in which narcolepsy experiments were first conducted was in dogs, which also have high identity scores compared with humans. These species can all be found in a phylogeny tree on the Phylogeny page. Because of the high homology between mice and human hcrtr1 DNA and protein, and because of the prevalence of using mice as a model organism, I would use mice if i were to conduct an experiment regarding the hcrtr1 gene and narcolepsy.
References:
(1) HomoloGene
(2) Wormbank
(3) Phenobank
(4) Chimpanzee picture link
(5) Dog picture link
(6) Cattle picture link
(7) Mouse picture link
(8) Rat picture link
(9) BLAST: Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402.
(2) Wormbank
(3) Phenobank
(4) Chimpanzee picture link
(5) Dog picture link
(6) Cattle picture link
(7) Mouse picture link
(8) Rat picture link
(9) BLAST: Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402.
Eric Suchy, Email: [email protected], last updated: May 15, 2010